Heat Map Panel¶
The Heat Map panel shows expression values for genes that are associated with selected nodes and edges.
You may select any number of nodes and/or edges in the network. Selecting an edge is equivalent to selecting the two gene set nodes that are connected to the edge.
Note
“Where are my expression values?”
If there are more than 50 expression values per gene then the Compress: Data Set Median option will be automatically enabled. Set Compress to –None– to see the original values. See below for more details.
Warning
There is a known issue on Mac where sometimes selecting a value in the Compress: combo box has no effect. The workaround is to click on the combo box with the mouse, hold down the mouse button, hover the mouse pointer over the option you want, and then release the button.
Toolbar¶

- Expression Legend
- Click on an expression value cell in the table to show the Expression Legend. It will not be visible until a value is selected.
- Shows the value range and the color gradient for the data set associated with the selected expression value. Note the value range may be different for different data sets.
- Genes
- All
- Shows all of the genes from all of the selected gene sets (union).
- Common
- Shows only genes that are common to all selected gene sets (intersection).
- All
- Expressions
- Values
- Shows the raw values from the expression file(s). Expression values are rounded to two decimal places.
- Row Norm
- Row normalizes the expression values. For each value in a row of expression the mean of the row is subtracted followed by division by the row’s standard deviation.
- Log
- Takes the log of each expression value.
- Values
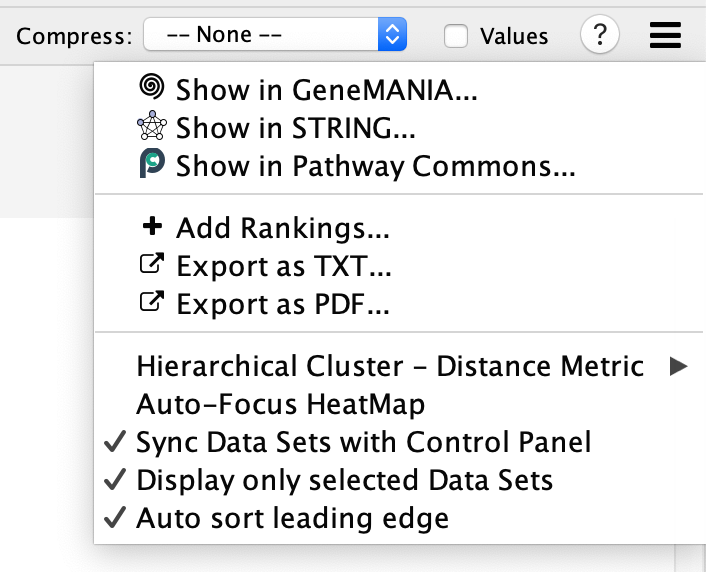
- Compress
- -None-
- Shows all of the expression values.
- Class: Median, Min, Max
- Only available if a class file was provided when the network was created.
- Shows a single column for each class where the value is the median, min or max of the values in the class.
- Data Set: Median, Min, Max
- Shows a single column for each data set where the value is the median, min or max of all the values.
- If the number of expressions per gene is greater than 50 then Compress: Median will be automatically enabled.
- -None-
- Values
- When disabled only the color gradients are shown. When enabled the numeric expression values are shown.
- Expression values are rounded to two decimal places.
- Menu button
- Opens the panel options menu.
Table¶
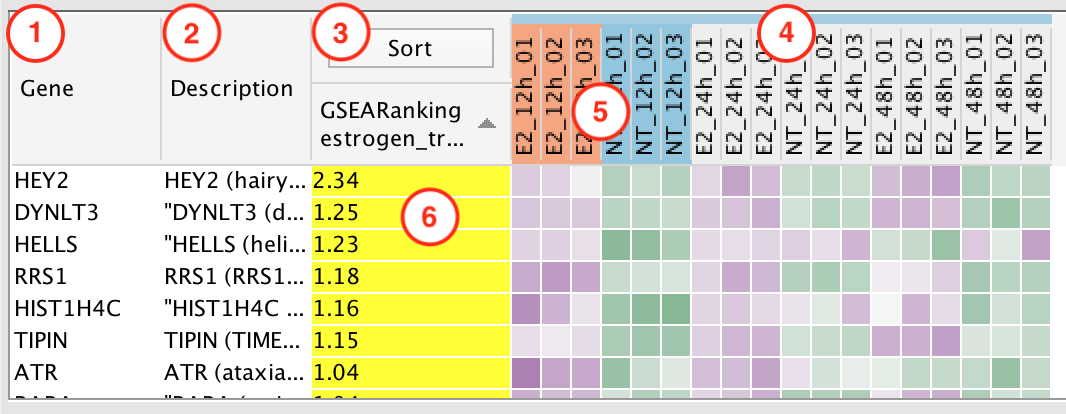
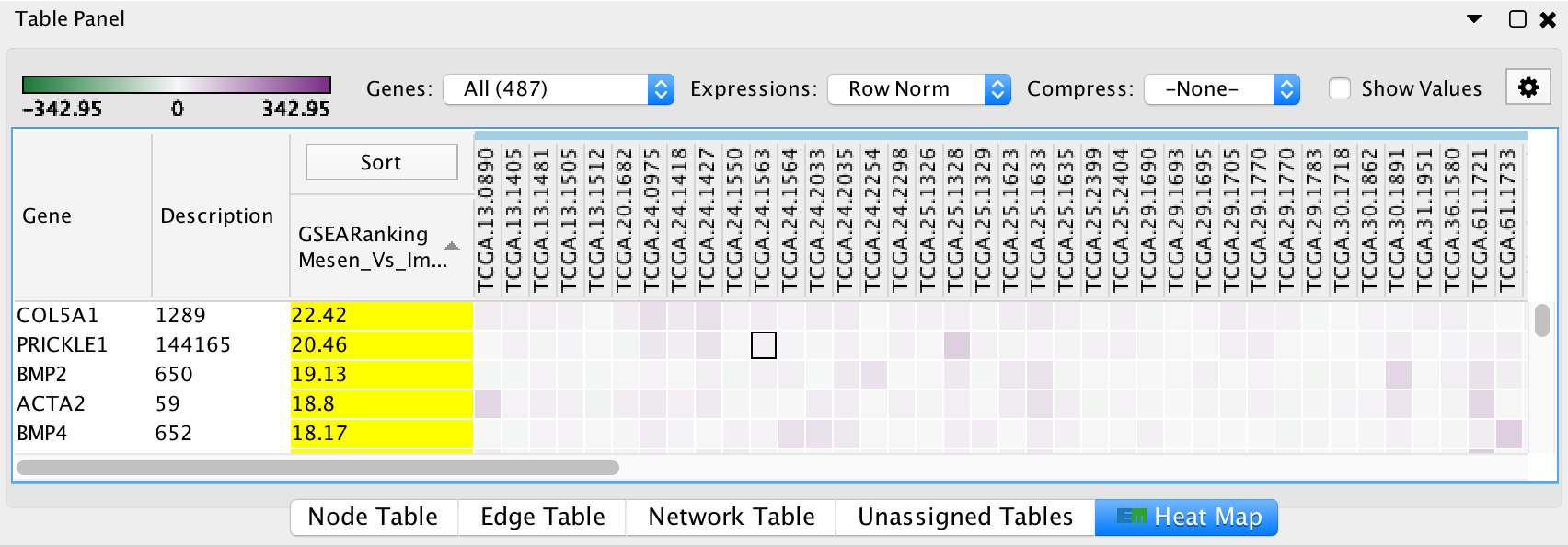
Click on any of the column headers to sort the table by that column.
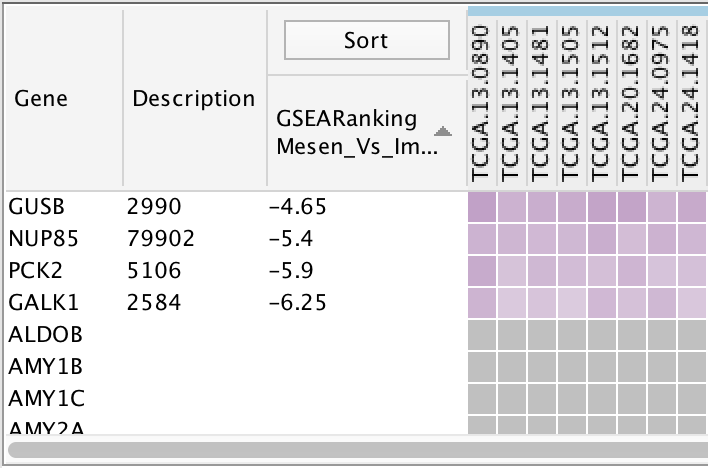
Gene Column
Description Column
Sort Column
- This column is used to sort by ranks or by hierarchical clustering.
- Click the Sort button to show a menu of ranking options.
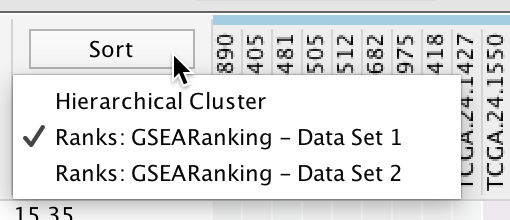
- If a data set has a rank file then the ranks will be listed in the menu.
- See Panel Menu below for details on how to load additional rank files.
- Hierarchical Clustering: Genes are clustered using a hierarchical clustering algorithm based on their expression values, the resulting hierarchy is then used to sort the genes.
Expression Columns
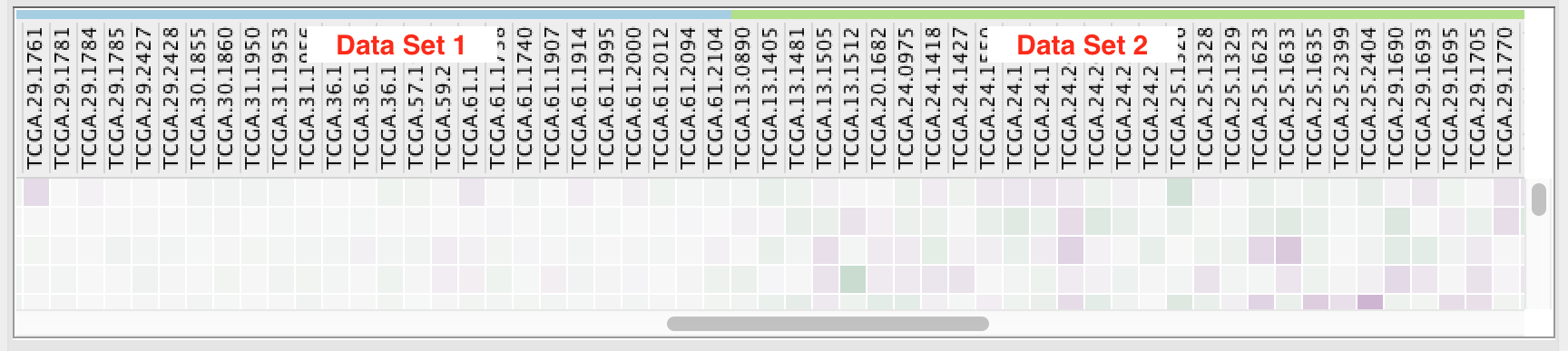
Shows expression values for each experiment.
If there is more than one data set and each data set has common expression values then the values will only be shown once.
If there are two or more data sets and they have different expression values then all the expression values are shown.
Genes that do not have expression data are shown in gray.
Phenotype Highlight
- The phenotypes that were entered in the Create EnrichmentMap Dialog are highlighted.
Leading Edge
- Genes that are part of the leading edge are highlighted in yellow.
- Available for GSEA results when a single gene set is selected.
- See below for more details.
GSEA Leading Edge¶
For every gene set that is tested for significance using GSEA there is a set of proteins in that gene set defined as the Leading Edge. According to GSEA the leading edge is:
“The subset of members that contribute most to the ES. For a positive ES, the leading edge subset is the set of members that appear in the ranked list prior to the peak score. For a negative ES, it is the set of members that appear subsequent to the peak score.”
In essence, the leading edge is the set of genes that contribute most to the enrichment of the gene set.
For Enrichment Map, leading edge information is extracted from the GSEA enrichment results files from the column denoted as Rank at Max. Rank at max is the rank of the gene where the ES score has the maximal value, i.e. the peak ES score. Everything with a better rank than the rank at max is part of the leading edge set.