Columns¶
EnrichmentMap creates several columns in the node and edge tables. They can be seen in the Node Table and Edge Table panels.
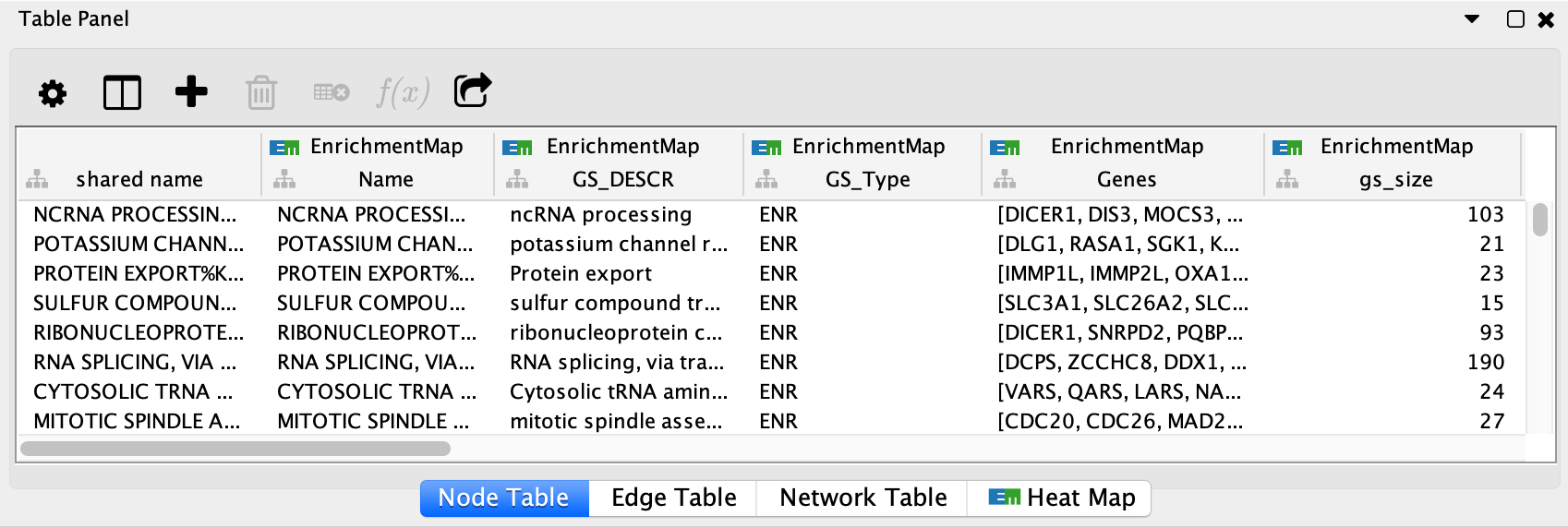
Columns created by EnrichmentMap have the following pattern:
EnrichmentMap::column_name (data_set_name)
EnrichmentMap::is the namespace prefix. This tells Cytoscape that the column is in the EnrichmentMap namespace.data_set_nameis used to differentiate between data sets. There will be one such column for each data set.
Note
Older versions of EnrichmentMap used the format EM1_column_name for column names.
Node Columns¶
- EnrichmentMap::Name
- The gene set name.
- EnrichmentMap::Formatted_name
- A wrapped version of the gene set name so it is easy to visualize.
- EnrichmentMap::GS_DESCR
- The gene set description (as specified in the second column of the gmt file).
- EnrichmentMap::Genes
- The list of genes that are part of this gene set.
- EnrichmentMap::gs_size
- Number of genes the union of the gene set across all data sets.
- EnrichmentMap::GS_Type
- Used by the visual style to discern between regular enrichment nodes and signature gene set nodes.
Additionally there are attributes created for each dataset:
- EnrichmentMap::pvalue (…)
- Gene set p-value, as specified in GSEA enrichment result file.
- EnrichmentMap::fdr_qvalue (…)
- Gene set q-value, as specified in GSEA enrichment result file.
- EnrichmentMap::Colouring (…)
- Enrichment map parameter calculated using the formula 1-pvalue multiplied by the sign of the ES score (if using GSEA mode) or the phenotype (if using the Generic mode)
GSEA specific attributes (these attributes are not populated when creating an enrichment map using the generic mode).
- EnrichmentMap::ES_dataset (…)
- Enrichment score, as specified in GSEA enrichment result file.
- EnrichmentMap::NES_dataset (…)
- Normalized Enrichment score, as specified in GSEA enrichment result file.
- EnrichmentMap::fwer_qvalue (…)
- Family-wise error score, as specified in GSEA enrichment result file.
Edge Columns¶
For each Enrichment map created the following attributes are created for each edge:
- EnrichmentMap::Data Set
- Contains the name of the data set that the edge is associated with, or ‘compound’ if the Combine edges across data sets option was selected when the network was created.
- EnrichmentMap::Overlap_size
- The number of genes associated with the overlap of the two gene sets that this edge connects.
- EnrichmentMap::Overlap_genes
- The names of the genes that are associated with the overlap of the two gene sets that this edge connects.
- EnrichmentMap::similarity_coefficient
- The calculated coefficient for this edge.